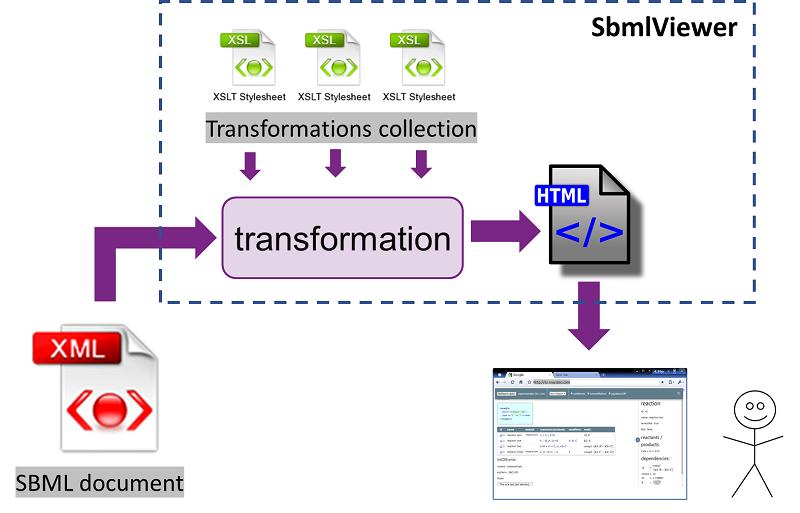
SbmlViewer is a tool for fast and easy reading of biological models written in SBML format. It allows looking through model components and equations even if you have no modeling software installed. It is as simple as reading a web page in your web browser.
SbmlViewer never uploads your files to the server and all transformations are performed locally in your browser, so it is safe and secure for your code.
SbmlViewer is open project located on GitHub. Any feedback is welcomed on Issues page.
SbmlViewer is helpful when:
- You need to share your model in human readable and compact format.
- You cannot install you favorite tool to read and check some SBML file right now.
- Your software cannot read some specific SBML versions or elements like
event,constraintorfunctionDefinitionetc. but the file includes them. - Your have got some errors importing SBML and you are trying to check the model code.
Quick start
For models from your computer:
- Open the page https://sv.insysbio.com/online
- Drug’n’drop your SBML or click on button “Choose file” and select your sbml model.
For models shared on web:
Some servers don’t allow to use shared files in this way. In that case download the file and use the method above.
- Copy the link of your shared model, for instance https://www.example.com/your_sbml_file.xml
- Add the external SBML path to SbmlViewer url followed by ? sign, like this:
https://sv.insysbio.com/online?https://www.example.com/external_sbml_file.xml - The full url above can be shared, sent or embedded to web page if you wish.
Examples
To look through example just try the links
- test 00001 from SBML Test Suite Database
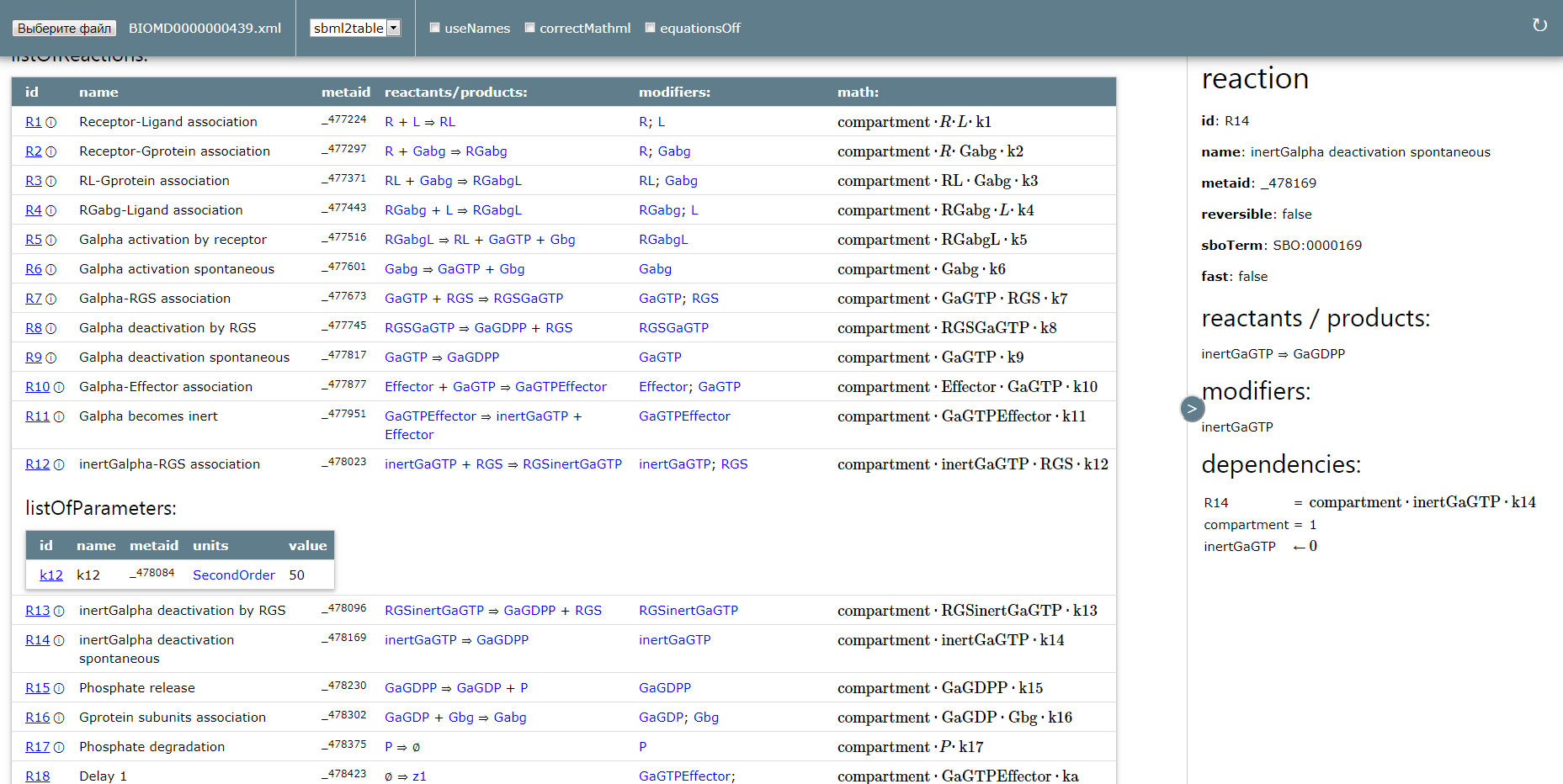
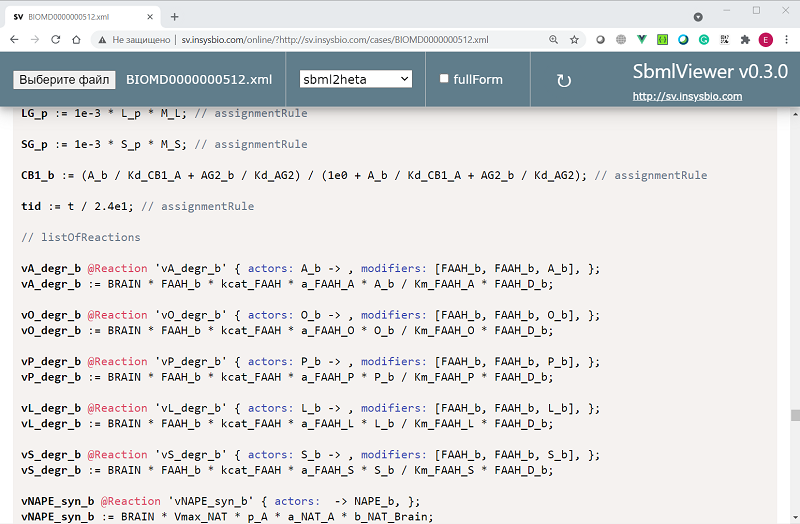
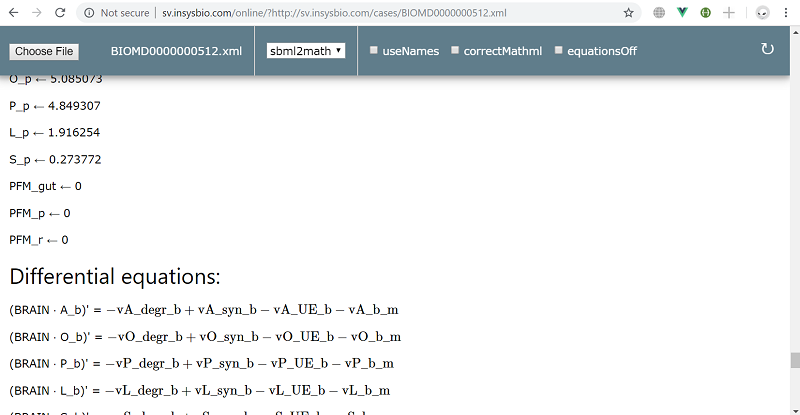
- BIOMD0000000512 from BioModels Database, model of the month in January 2016
- BIOMD0000000588 from BioModels Database, model of the month in February 2017
- BIOMD0000000439 from BioModels Database, model of the month in October 2016
Features list
- reading SBML Level 2 Version 1-5
- reading SBML Level 3 Version 1-2
- transformation to Heta code using sbml2heta or sbml3heta
- transformation to the Antimony format antimony2heta or antimony3heta
- creating tabular view of model with: sbml2table or sbml3table transformation
- summary generation for a chosen element with sbml2element transformation
- generation of ODE system based on your model: sbml2math or sbml3math transformation
- supports specific SBML format generated by simbiology by transforming MathML and using name instead of id, see options
- Support nice printing of the whole document:
Ctrl + P
Browser support
Tested on:
- Google Chrome
- Opera
- Mozilla Firefox
- MS Internet Explorer
- MS Edge
- Safari
License
Apache 2.0
Team
About SBML
SBML is a free and open interchange format for computer models of biological processes maintained by SBML community. It is used in many modeling application and can store model structure, math and annotation of elements. SBML is XML based format and designed basically for machine reading and writing.
